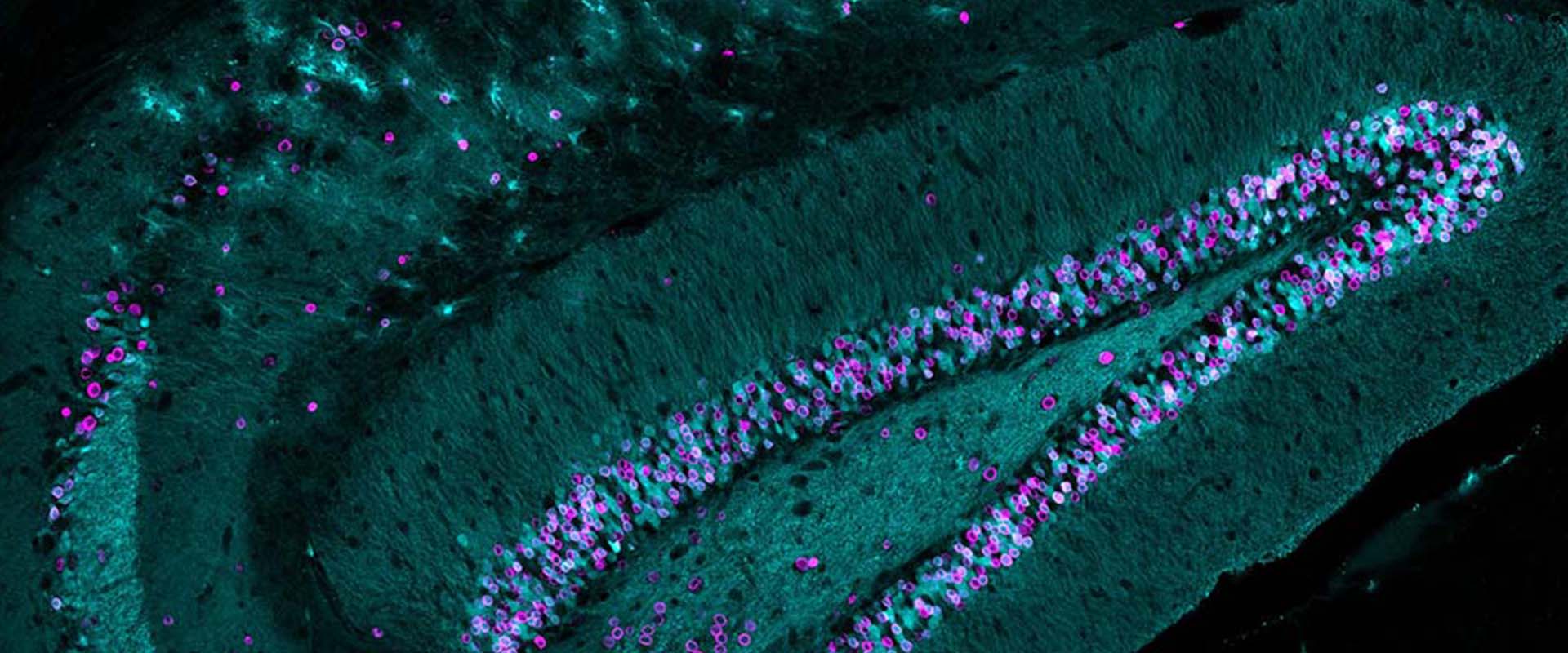
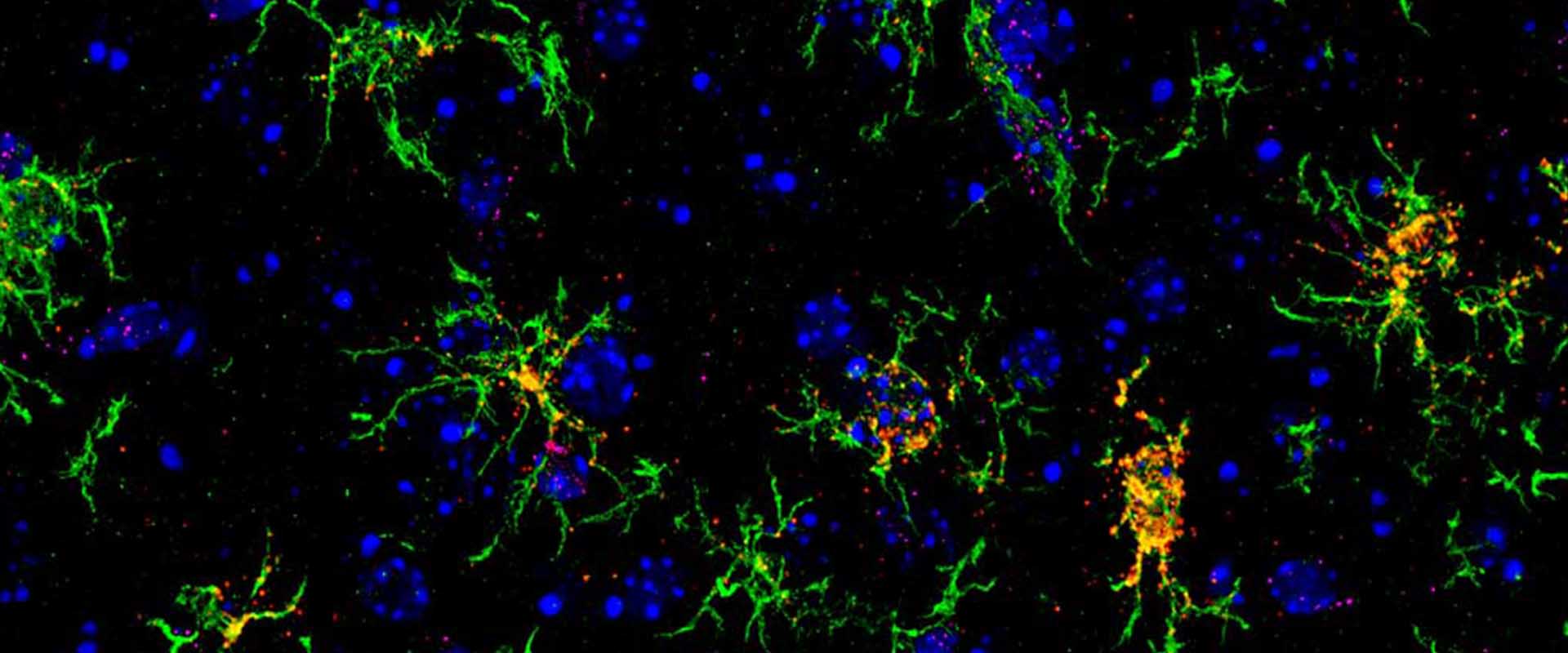
Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibiton
We and others have previously shown that the epigenetic drugs histone deacetylase inhibitors (HDACi) potentiate memory and synaptic plasticity and ameliorate cognitive deficits and degeneration in animal models for different neuropsychiatric conditions such us Rubinstein-Taiby syndrome and Alzheimer’s and Huntington’s disease. However, the impact of these drugs on neural histone acetylation and gene expression at the genomic level, and the molecular mechanisms that underlie their specificity and beneficial effects in neural tissue remain obscure. In the current study, we mapped four relevant histone marks (H3K4me3, AcH3K9,14, AcH4K12 and pan-AcH2B) in hippocampal chromatin and investigated at the whole-genome level the impact of HDAC inhibition on acetylation profiles and basal and activity-driven gene expression. The combination of differential expression (microarrays) and histone acetylation (ChIP-seq) screenings demonstrated that HDAC inhibition caused a dramatic hyperacetylation of the different histones that was largely restricted to active loci were already labelec with acetylation marks at the basal state. In addition, the restricted changes in hippocampal gene expression associated with TSA-induced histone hyperacetylation and its negligible impact on the transient transcriptional response to neuronal activation led us to hypothesize that the beneficial effects of HDACi in neural tissue may be mediated by histone acetylation-independent activation of gene programs downstream of transcription factors that are also substrate of HDAC enzymes. Our results illuminate both the relationship between hippocampal gene expression and histone acetylation and the mechanism of action of these important neuropsychiatric drugs.

 Español
Español